Disclosing the hidden layer of splicing-effective mutations in cancer
August 2018 .Frankfurt scientists together with scientists from the IMB Mainz decode the alternative splicing of the proto-oncogene RON with a high-throughput mutagenesis screen.
Mutations causing aberrant splicing are frequently implicated in human diseases including cancer. Splicing requires tight control of trans-acting factors that recognise cis-regulatory elements in the RNA sequence. However, the position and function of most cis-regulatory elements remain unknown, hindering the interpretation of disease-associated mutations and resulting splicing changes
Researchers from BMLS at Goethe University Frankfurt and IMB in Mainz established a high-throughput screen of randomly mutated minigenes to decode the cis-regulatory landscape of selected splicing decisions. As a prototype example, they screened the cancer-relevant alternative exon 11 in the proto-oncogene MST1R (RON) encoding for a receptor tyrosine kinase. Skipping of RON exon 11 results in the pathological isoform RONΔ165 results that promotes tumour invasiveness and is frequently upregulated in solid tumours.
Mathematical modelling of splicing kinetics enables the scientists to identify more than 1,000 mutations affecting RON exon 11 skipping. Importantly, the measured effects correlate with RON alternative splicing in cancer patients bearing the same mutations. Moreover, they highlight the RNA-binding protein heterogeneous nuclear ribonucleoprotein H (HNRNPH) as a key regulator of RON splicing in healthy tissues and cancer. Using iCLIP and synergy analysis, the researchers pinpoint the functionally most relevant HNRNPH binding sites and demonstrate how cooperative HNRNPH binding facilitates a splicing switch of RON exon 11. Their results thereby offer insights into splicing regulation and the impact of mutations on alternative splicing in cancer.
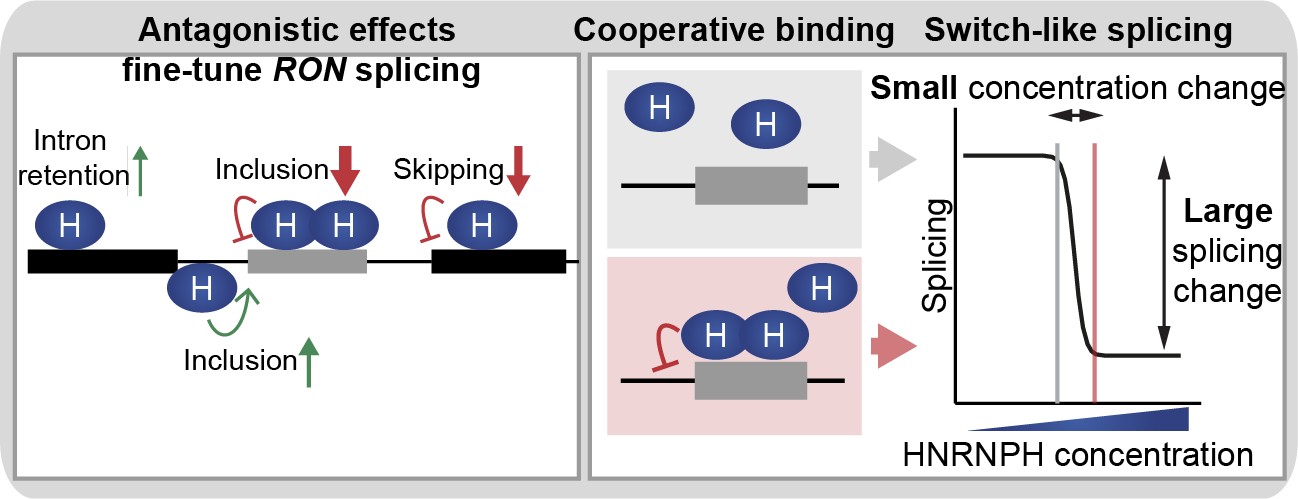
The RNA-binding protein HNRNPH (heterogeneous nuclear ribonucleoprotein H) HNRNPH acts as key regulator of alternative splicing in the proto-oncogene RON. To fine-tune the regulation, HNRNPH recognises multiple cis-regulatory elements in the RNA sequence in a cooperative fashion.
Schematic model summarises position-dependent effects of HNRNPH on RON exon 11, indicating most strongly effected splice isoform for each HNRNPH binding site (left panel). Multiple interdependent HNRNPH binding sites within RON exon 11 exert strong cooperative control on the alternative exon, resulting in a splicing switch upon small changes in HNRNPH abundance (right panel)
Figure taken from Braun et al., Nature Communications
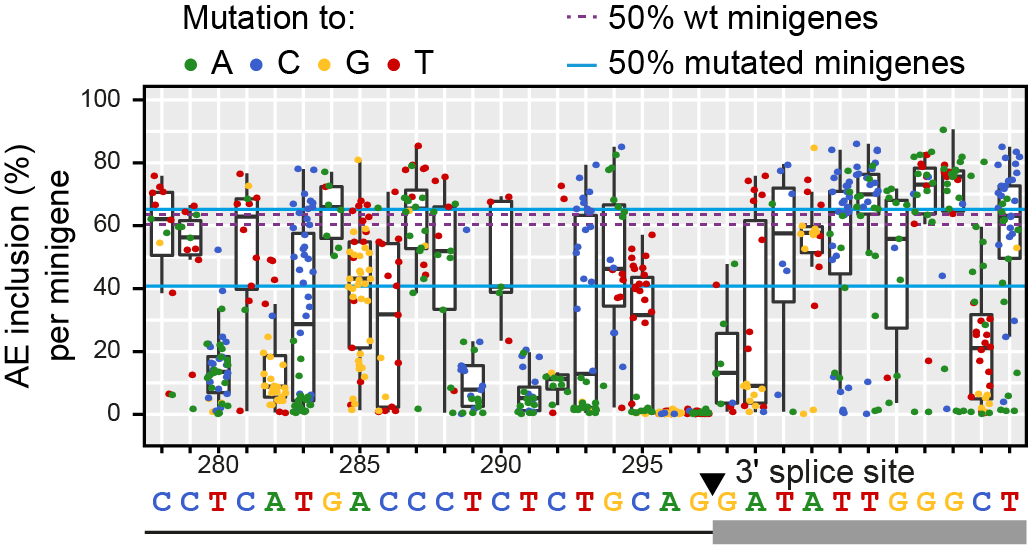
Mutational landscape around the 3’ splice site of alternative exon 11 in RON. Boxplot of alternative exon (AE) inclusion frequencies (in percent over all splice isoforms) in human HEK293T cells for all minigenes with a mutation at the indicated position (x-axis). Each position was mutated in 28 different minigenes on average, providing sufficient replication for reliable splicing estimates. Colours illustrate inserted nucleobase (see legend). Blue and purple lines indicate interquartile range (IQR) of AE inclusion frequencies for all mutated and wild type (wt) minigenes (i.e. without mutations), respectively. Sequence of wt RON minigene given below.
Figure taken from Braun et al., Nature Communications
Contacts:
Dr. Kathi Zarnack
Buchmann Institute for Molecular Life Sciences
Goethe University Frankfurt
kathi.zarnack(at)bmls.de
Dr. Julian König
Institute of Molecular Biology (IMB), Mainz
j.koenig(at)imb-mainz.de
Dr. Stefan Legewie
Institute of Molecular Biology (IMB), Mainz
s.legewie(at)imb-mainz.de
Publication:
Braun, S*, Enculescu, M*, Setty, ST*, Cortés-López, M, de Almeida, BP, Sutandy, FXR, Schulz, L, Busch, A, Seiler, M, Ebersberger, S, Barbosa-Morais, NL, Legewie, S$, König, J$, Zarnack, K$ (2018) Decoding extensive regulation of RON alternative splicing with a high-throughput mutagenesis screen. Nat Commun, doi: 10.1038/s41467-018-05748-7
(* Shared first authorship; $ corresponding authors)