Projects in Cellular and Molecular Neurobiology
- Project 1
- Project 1-1
- Project 1-2
- Project 1-3
- Project 2
- Project 2-1
- Project 2-2
- Project 3
- Project 3-1
- Project 3-2
- Project 3-3
- Project 3-4
- Methods I
- Methods II
- nAChRs
Functional Connectomics: How does the nervous system affect behavior?
Caenorhabditis elegans has an anatomically well-defined nervous system of just 302 neurons, whose synaptic connections have been mapped precisely by serial electron microscopy.
Knowledge of the cellular connections, however, is only one key to understanding how the circuits work, and only limited insight into function can be deduced from the pure connectome. This is because this dataset does not contain information about the ‘sign’ of individual synapses, the transmitters and postsynaptic receptors used, as well as the synaptic weight and dynamic signalling.
Furthermore, neuronal networks of the worm are more complex than was assumed, because of the adoption of different functional states depending on context and/or the presence of certain neuromodulators. Such brain states involve that anatomical synapses are being switched to active or inactive states, such that several different functional connectomes are overlaid on one anatomical, hard-wired network.

Thus, to enable understanding the neuronal network of the nematode, we use functional connectomics. This approach takes the anatomical connectome, and tries to work out the individual connections between pairs of cells or groups of cells in a functional manner.

Optogenetic dissection of food-motivated behavior
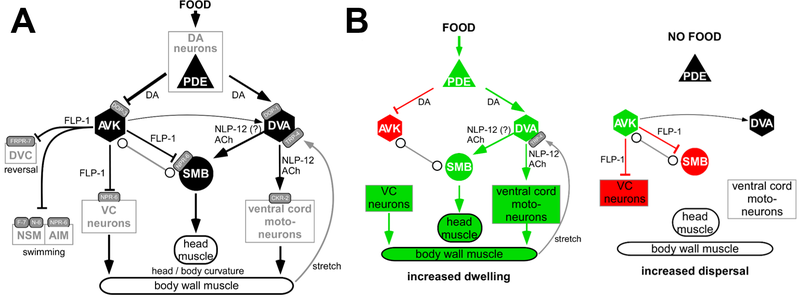
We analyzed a network of neurons that regulates 'strategic' navigational behavior of the animal in the presence or absence of food (bacteria). On a food patch, animals slowly forage, a behavior termed dwelling. If food is removed, they initially stay in the vicinity to search food locally, but if unsuccessful for a couple of minutes, they switch to so-called dispersal behavior, characterized by long straight runs, to search for food elsewhere. This behavioral switch is orchestrated, among other cell types, by a network described below. Dopaminergic sensory PDE neurons, that detect the presence of food outside the body, convey this signal to two types of interneurons, AVK and DVA, by two different dopamine receptors, DOP-1 and DOP-3, that are excitatory or inhibitory, respectively.
Oranth, A., et al., Food Sensation Modulates Locomotion by Dopamine and Neuropeptide Signaling in a Distributed Neuronal Network. Neuron, 2018. 100(6): p. 1414-1428 e10
The AS class of motor neurons coordinate locomotion
Many aspects of the neuronal control of locomotion in C. elegans are not yet understood. We could recently fill an important gap in the understanding of how the worms undulatory wave-like movement is controlled. Using optogenetic, behavioural and imaging analyses, we studied the remaining ‘unknown’ class of AS cholinergic motor neurons We found that these neurons generate asymmetric muscle activation, enabling navigation, and contribute to coordination of dorso-ventral undulation as well as anterio-posterior bending wave propagation.
Tolstenkov O, Van der Auwera P, Steuer Costa W, Bazhanova O, Gemeinhard T, Bergs ACF, Gottschalk A. (2018) Functionally asymmetric motor neurons contribute to coordinating locomotion of Caenorhabditis elegans. eLife 7: e34997: PDF

Optogenetic analysis of a nociceptive neuronal network
Neuronal networks underlie behavior in every animal. C. elegans has a very simple and anatomically well-defined nervous system, allowing to decipher how neurons generate a particular behavior, and how they interact in networks. Such approaches are largely supported by optogenetic methods, as they allow to acutely stimulate, inhibit or otherwise modify neuronal function in live, freely behaving animals. We used these methods to decipher a nociceptive neuronal network of C. elegans, which involves the multidendritic neurons FLP and PVD, that cover head and body of the worm with fine processes and detect harsh touch, among other modalities. We investigated how these neurons interact with command interneurons to evoke backward or forward escape responses. The PVD-evoked forward escape behavior was further used to analyze the cell-autonomous function of genes specifically expressed in PVD. We found that the TRPM channel GTL-1 acts as a general amplifier of signals within PVD, and that the DEG/ENaC ASIC-1 potentiates the signal output of PVD and might extend its dynamic range.
Husson S, Steuer Costa W, Wabnig S, Stirman JN, Watson JD, Spencer WC, Akerboom J, Looger LL, Treinin M, Miller III DM, Lu H and Gottschalk A. (2012) Optogenetic analysis of a nociceptor neuron and network reveals ion channels acting downstream of primary sensors. Current Biology 22: 743-752.
Collaboration: David Miller III (Vanderbilt University), Hang Lu (Georgia Tech), Millet Treinin (Hebrew University)

Mechanisms of synaptic transmission and recycling
My lab studies molecular and cellular aspects of synaptic transmission at chemical synapses, specifically the neuromuscular junction. We use optogenetic stimulation of neurons, behavioral analysis, electrophysiology, imaging and electron microscopy. This way, also aspects of the ultrastrucutral changes evoked by neuronal activity can be studied, acutely, following or during strong optogenetic stimulation.


Neuropeptides regulate synaptic transmission and quantal content
Neurons store neurotransmitters in synaptic vesicles (SVs). Electrical activation of the neuron leads to fusion of the vesicles with the outer envelope of the neuron, and thus to release of the transmitters. This usually occurs in discrete quantities, determined by the volume and the filling state of the vesicles. Neurons can fuse vesicles at a lower or higher rate, thus releasing more or less quanta of transmitter per time - a molecular ‘gas pedal’ of the neuron. However, it could also be useful to alter the filling state of the vesicles, e.g. adapted to certain situations which require a stronger neural activity. To remain in the above analogy, this could be interpreted as a 'gear shift': with the same amount of gas, a higher speed can be achieved in a higher gear. We identified a mechanism of this gear shift in cholinergic motor neurons, which depends on neuropeptide signaling. Neuropeptides are released in response to (optogenetic) cAMP increase, and induce mobilization of SVs. However, they also cause additional filling of SVs with acetylcholine within a few seconds. This is likely to occur on existing SVs, is visible at the ultrastructural level, and leads to observably higher amplitude of postsynaptic currents.
Steuer Costa W, Yu S-c, Liewald JF, Gottschalk A*. (2017) Fast cAMP modulation of neurotransmission via neuropeptide signals and vesicle loading. Curr. Biol. 27: 495-507.

Synaptic vesicle recycling
Synaptic vesicles (SVs) need to be recycled locally, in the synaptic terminal, to sustain high activity of the neuronal signaling to downstream cells. We analyzed in a number of studies the mechanisms of SV recycling, developing optogenetic methods ('OptIoN' - optogenetic investigation of neurotransmission) for stimulation of neurons, combined with electrophysiology, behavioral analysis and electron microscopy
Liewald JF, Brauner M, Stephens GJ, Bouhours M, Schultheis C, Zhen M, Gottschalk A. Optogenetic analysis of synaptic function. Nat. Meth. (2008) 5: 895-902.
Kittelmann M, Liewald JF, Hegermann J, Schultheis C, Brauner M, Steuer Costa W, Wabnig S, Eimer S*, and Gottschalk A.* (2013) In vivo synaptic recovery following optogenetic hyperstimulation. PNAS 110: E3007-E3016
Wabnig S, Liewald JF, Yu S-c, Gottschalk A (2015) High-throughput all-optical analysis of synaptic transmission and synaptic vesicle recycling in Caenorhabditis elegans. PLoS ONE 10(8): e0135584. doi:10.1371/journal.pone.0135584 PDF
Steuer Costa W, Yu S-c, Liewald JF, Gottschalk A. (2017) Fast cAMP modulation of neurotransmission via neuropeptide signals and vesicle loading. Curr. Biol. 27: 495-507
Yu S-c*, Jánosi B*, Liewald J, Wabnig S, Gottschalk A. (2018) Endophilin A and B join forces with clathrin to mediate synaptic vesicle recycling in Caenorhabditis elegans. Front Molec Neurosci 11: 196.
Optogenetic tool development
We have established the optogenetic toolbox for the nematode C. elegans. This includes rhodopsin-based tools for de- and hyperpolarization, cyclic nucleotide manipulation (cAMP, cGMP), imaging of Ca2+ and cGMP, rhodopsin-based tools for voltage imaging, and tools for protein degradation.

Rhodopsins for actuation
Channelrhodopsin, Halorhodopsin, Anion Channelrhodopsins, Full-length Inverted Channelrhodopsin, color shifted Channelrhodopsins, Step-function Opsins, Proton pumps, etc.
Nagel et al. (2005) Curr. Biol. 15:2279-2284.
Zhang et al. (2007) Nature 446:633-9.
Schultheis et al. (2011) PLoS ONE 6: e18766.
Husson et al. (2012) PLoS ONE 7: e40937.
Erbguth et al. (2012) PLoS ONE 7: e46827.
AzimiHashemi et al. (2014) Nat. Communic. 5: 5810.
Bergs et al. (2018) PLoS ONE 13: e0191802.
Brown et al. (2018) Cell 175: 1131-1140.
Collaborations with G. Nagel (Würzburg), E. Bamberg (Frankfurt), P. Hegemann (Berlin), K. Deisseroth (Stanford), A. Karpova (Janelia Farm)
Tools for voltage and Ca2+ imaging
We have established rhodopsin based tools for voltage imaging in muscles and neurons, as well as spectrally compatible genetically encoded Ca2+ indicators for combination with rhodopsin actuators.
Azimi Hashemi N*, Bergs ACF*, Schüler C, Scheiwe AR, Steuer Costa W, Bach M, Liewald JF, Gottschalk A. (2019) Rhodopsin-based voltage imaging tools for use in muscles and neurons of Caenorhabditis elegans. PNAS https://doi.org/10.1073/pnas.1902443116
Akerboom, J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolö J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schüler C, Chen T-W, Sarkisyan, KS, Marvin JS, Bargmann, CI, Kim DK, Kügler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER and Looger LL. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging in combination with optogenetics. Front. Molec. Neurosci. 6: 2
Collaboration with L. Looger (Janelia Farm)

Electron Microscopy of Photostimulated Chemical Synapses
We have established optogenetic tools for the generation of cAMP and cGMP, based on BLUF domain proteins and rhodopsins, as well as tools for imaging of cGMP.
Weissenberger et al. (2001) J. Neurochem. 116(4):616-25.
Gao et al. (2015) Nat. Communic. 6: 8046. PDF
Steuer Costa et al. (2017) Curr. Biol. 27: 495-507
Woldemariam et al. (2019) Genetics https://doi.org/10.1534/genetics.119.302392
Collaborations with G. Nagel (Würzburg), N. L'Etoile (San Francisco)
Tools for protein degradation
We have established optogenetic tools for degradation of any protein of interest (photosensitive degron), as well as a tool for specific photo-cleavage of synaptobrevin, leading to blockade of transmitter release.
Hermann A, Liewald JF, Gottschalk A. (2015) A photosensitive degron enables acute light-induced protein degradation in the nervous system. Curr. Biol. 25: R733–R752
Liu Q, Sinnen BL, Boxer EE, Schneider MW, Grybko MJ, Buchta WC, Gibson ES, Wysoczynski CL, Ford CP, Gottschalk A, Aoto J, Tucker CL, Kennedy MJ. (2019) A photoactivatable botulinum toxin for inducible control of neurotransmission. Neuron 101: 863-875.
Collaborations with C. Taxis (Marburg), C. Tucker (Aurora, Colorado)


Multicolor, selective photostimulation of neurons in freely moving animals
Specific photostimulation or inhibition of specific or even single neurons in C. elegans using microbial rhodopsins requires specific expression in just this cell, such that it can be activated by wide-field illumination. As such specific expression is hard to achieve, one could also restrict the light to just the region of the body that contains the neuron of interest. If this can be done in freely moving animals, one could influence the behavior in an acute fashion, while it occurs. Furthermore, multiple rhodopsins that are responsive to different colors of light could be combined in cells of a circuit, to probe different nodes in their effects on the behavior. Such multicolor illumination of distinct regions of the animal was achieved by our collaborators Jeff Stirman and Hang Lu (Georgia Tech). They devised a system that tracks an individual animal, and then projects a predefined multicolor light pattern on the animal, faithfully, while it is moving. We designed key proof of principle experiments to develop and test this system, and are now using it routinely in our lab.

In vivo labeling of cell surface exposed proteins in C. elegans
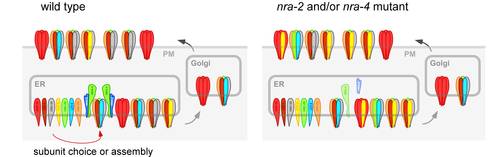
The most frequently used method to study expression patterns of proteins in C. elegans is to tag the protein of interest with GFP and express such a transgene in vivo. Especially for proteins expressed in the nervous system (for example neurotransmitter receptors), it is often desireable to visualize only those proteins that are inserted in the plasma membrane and thus exposed on the cell surface in a functional manner (for example as part of post-synaptic receptor clusters). However, GFP-tagged transgenes often hamper such investigations, because the protein is also found intracellularly and its fluorescence masks the signal from proteins that are inserted in the plasma membrane. Also, with GFP one cannot be sure if a spot of fluorescence belongs to proteins that are inserted in the plasma membrane, or to proteins that are present in vesicles close to, but not as part of the plasma membrane. We thus developed a method for specific immunolabeling of cell-surface exposed proteins and other extracellular epitopes. The protein of choice, for example a nAChR subunit, is expressed with a short peptide epitope tag (or GFP) that is placed extracellularly, if the protein is correctly inserted in the plasma membrane.

Fluorophore-labeled antibodies specific for this epitope tag are then injected into the body cavity of C. elegans, from where they get access to those epitopes on cell surfaces. Excess antibody is filtered from the extracellular fluid by the six scavenger cells, that constantly take up this fluid to remove metabolic breakdown products, thus leaving a clean staining patterns, essentially without background fluorescence. Alternatively, one can also use natural ligands of the protein of interest, labeled with a fluorophore, to label this protein, for example α-bungarotoxin, that binds to the nAChR. Examples of such staining have been published in McKay et al., 2004, Genetics 166, p. 161-9 and Zheng et al., 2004, Nature 427 pp 451-7, Gottschalk and Schafer, 2006 J. Neurosci. Meth. in press.

Multicolor, selective photostimulation of neurons in freely moving animals
Proteins that act as regulators of nAChR function, for example through direct post-translational modification (like protein kinases), must physically interact with nAChR subunits. Thus they can be identified through biochemical co-purification with nAChRs. We have purified the so-called levamisole-sensitive nAChR from C. elegans, which is expressed in both muscles and many neurons (including motorneurons). To do this, we used the tandem affinity purification (TAP; see below). Proteins that were co-purified with TAP-tagged levamisole receptor subunits were identified in the whole sample, using 2D-HPLC of trypsin-digested peptides and nanoelectrospray-tandem mass spectrometry. In addition to abundant nAChR subunits, we could identify proteins that interact with the nAChR with very low affinity, and thus were highly underrepresented in the sample. To distinguish those from co-isolated contaminating proteins, we use a functional assay. Each of the proteins is depleted in vivo using doublestranded RNA interference (RNAi), and the responses of these animals to nicotine are then assayed. Thus we could identify a number of proteins whose depletion either caused nicotine resistance or hypersensitivity, making them prime candidates for factors affecting nAChR biology. We are characterizing these proteins in detail, and two of these proteins were found to act in the ER to influence the subunit composition of nAChRs. Gottschalk et al., 2005, EMBO J. 24, p 2566-2578. Almedom RB, Liewald J, Hernando G, Schultheis C, Rayes D, Pan J, Schedletzky T, Hutter H, Bouzat C, Gottschalk A. 2009. EMBO J. 28:2636-49.